Patient suffers severe amnesia but musical memory remains intact
Posted by Acubiz | BlogTogether with his team, Prof. Christoph Ploner, director of the Department of Neurology at the Virchow campus, examined a professional cellist who suffered from encephalitis caused by a herpes virus. As a result of the inflammation, the patient developed serious disturbances in memory.
Both his memory for the past (retrograde amnesia), as well as the acquisition of new information (anterograde amnesia) were affected. Whereas the patient was unable to recount any events from his private or professional life, or remember any of his friends or relatives, he retained a completely intact musical memory. Furthermore, he was still able to sight-read and play the cello.
For the systematic examination of his musical memory, Dr. Carsten Finke, Nazli Esfahani and Prof. Christoph Ploner developed various tests that take the beginning of his amnesia into account. In comparison to amateur musicians and professional musicians from the Berlin Philharmonic, the patient showed a normal musical memory in all tests. He not only remembered music pieces from the past, but was also able to retain music he had never heard before.
“The findings show that musical memory is organized at least partially independent of the hippocampus, a brain structure that is central to memory formation,” says Carsten Finke, the primary author of the study. “It is possible that the enormous significance of music throughout all times and in all cultures contributed to the development of an independent memory for music.”
Carsten Finke and his colleagues hope that the intact musical memory in patients with amnesia can be used to stimulate other memory content. In this way, perhaps a particular melody can be connected to a person or an everyday task, for example taking medicine.
New study finds promising results for MERS treatment
Posted by Acubiz | BlogApproach uses genetically modified cows to manufacture MERS-fighting antibodies
In a new study, University of Maryland School of Medicine researchers have had promising results with a new treatment for Middle East Respiratory Syndrome (MERS). The study, published in the journal Science Translational Medicine, found a new treatment that protected mice from MERS infection.
The treatment — an antibody that blocks the MERS virus — was produced by cows that had been genetically modified to mimic certain aspects of the human immune system. These cows were given a new MERS vaccine that led to production of anti-MERS antibodies in large quantities. These antibodies were then purified to produce the therapeutic that was tested in the MERS-infected mice.
Last year, a South Korean epidemic of MERS killed more than 30 people. Overall, MERS has killed nearly 600 people since it was first discovered four years ago in Saudi Arabia. The South Korean outbreak, which began when a traveler returned from Saudi Arabia, infected hundreds of people there.
The research is a partnership between the University of Maryland School of Medicine (UM SOM), SAB Biotherapeutics (SAB), Novavax and the Naval Medical Research Center. The researchers tested the treatment for MERS, a disease that can cause severe respiratory symptoms and has a death rate of 40 percent.
“These results are very promising,” says one of the lead researchers on the study, Matthew B. Frieman, PhD, an Associate Professor of Microbiology and Immunology at UM SOM. “This is important not only because it gives us a potential way to attack MERS, but also because it provides evidence that using these transgenic cows can rapidly produce therapeutics.”
SAB, a biopharmaceutical company based in Sioux Falls, South Dakota, provided the genetically modified cows, a technology that it invented. Novavax, a vaccine biotech company based in Gaithersburg, provided the vaccine that triggered the antibody production in the cows.
“Through this collaborative team, we’ve brought together the top talent of the scientific community, global health experts and novel technologies to demonstrate the efficacy, safety, and responsiveness of our human antibody therapeutic,” said Dr. Eddie Sullivan, PhD, President and CEO of SAB Biotherapeutics, Inc. “As we complete successful studies targeting various diseases, we’re realizing the potential broad application and significance of the our platform in addressing these global health threats.”
The next step, which will occur in the next three to six months, will be a human clinical trial to test the safety of the therapeutic. If that works, a Phase 2 trial will follow, to test whether it is effective for use in humans, in emergency situations.
MERS was first discovered in 2012 in Saudi Arabia. It appears that the disease spread to humans from camels, who may themselves been infected by bats. Research has shown that it is similar to Severe acute respiratory syndrome (SARS), which emerged in 2003 and resulted in over 8000 infections, killing 10% of those infected. Both are caused by Coronaviruses, both cause serious respiratory problems, and both are often fatal.
“Prof. Frieman’s work is a fantastic example of how the school is partnering with private industry to break new ground fighting disease,” said UM SOM Dean E. Albert Reece, MD, PhD, MBA, who is also the vice president for Medical Affairs, University of Maryland, and the John Z. and Akiko K. Bowers Distinguished Professor. “This work provides a great model for how we can respond rapidly to emerging diseases that threaten health around the world.”
The RNA alphabet: The key role played by hmC
Posted by Acubiz | BlogLed by François Fuks from the ULB’s Laboratory of Cancer Epigenetics and the ULB-Cancer Research Center (U-CRC), researchers have revealed for the first time the key role played by one of the RNA letters, hmC or hydroxymethylation. Their discovery will help us better understand such diseases as cancer.
DNA is made up of 4 letters or nucleotides (A, T, G, C), the sequence of which determines our genome. We know that there is a fifth letter completing the genome: DNA methylation (mC). This helps in cell specialisation through controlling the expression of certain genes. When these genes are not correctly methylated, there is a risk of their expression being altered, leading to the emergence of such diseases as cancer. Therapies correcting such methylation faults are already being used to treat cancer.
RNA is the other molecule of life. For several years now, we have been witnessing a paradigm change, with RNA now seen as being just as important as DNA in understanding the book of life. Indeed, it would seem that RNA is not just an intermediary between DNA and protein, but is capable of explaining several major mysteries in the study of life, such as the origin of life and the “junk DNA” paradox.
Through putting the spotlight on RNA, a completely new research path emerges: the complex RNA alphabet (or RNA epigenetics). Just as with DNA, in addition to the 4 well-known letters (A, U, G, C), there are further letters defining the chemical properties of RNA. Yet the importance of RNA epigenetics for cell development remains unexplored …
The recent work of the team led by Prof. François Fuks, head of the ULB Faculty of Medicine’s Laboratory of Cancer Epigenetics and of the ULB Cancer Research Center, U-CRC, has revealed the key role played by one of these RNA letters, hydroxymethylation (hmC).
Using fruit flies, one of the most common model organisms in biology, the ULB researchers have shown that hmC promotes the translation of RNA into proteins. In addition, following the introduction of a new high-performance sequencing technology, they have fully mapped the epigenetics of hmC. Last but not least, François Fuks and his colleagues have demonstrated the essential role played by hmC in cell development: when hmC production was impeded, the flies died.

Transgenic sweet corn no more susceptible to Goss’s wilt disease
Posted by Acubiz | BlogExperiment showed transgenic sweet corn was not more susceptible to Goss’s wilt disease when treated with glyphosate.
Credit: Marty Williams
Transgenic crops expressing resistance to the herbicide glyphosate (GR) have been commercialized and planted widely across the U.S. for two decades. The majority of transgenic corn (Bt) also has been engineered to produce toxins effective against certain corn insect pests. In recent years, claims have been made that glyphosate and transgenic traits result in corn plants that are more susceptible to crop diseases.
Such claims have linked the rise in occurrence of corn diseases like Goss’s wilt, which causes leaf blight and systemic wilt, to the adoption of transgenic corn across the U.S. However, a new study from the USDA-Agricultural Research Service (ARS) provides empirical evidence showing no increase in disease susceptibility in transgenic sweet corn treated with glyphosate.
“Results showed glyphosate use and transgenic traits were not factors in disease susceptibility,” says Martin Williams, a USDA-ARS ecologist and University of Illinois crop scientist.
The team tested a fresh-market sweet corn hybrid varying in the absence or presence of the GR+Bt transgenes; Passion and Passion II, respectively. A subset of both sweet corn lines were inoculated with the bacterium that causes Goss’s wilt before or after a label-standard glyphosate application. Passion was not treated with the herbicide because it does not have the transgene that is essential for plant survival in the presence of glyphosate.
Approximately one-half of the inoculated plants developed symptoms of Goss’s wilt, regardless of the presence or absence of transgenic traits. Moreover, the timing of disease inoculation with respect to glyphosate application did not influence Goss’s wilt incidence or severity.
Williams notes, “The only factor affecting Goss’s wilt incidence was whether or not plants were inoculated. We found no evidence of differential susceptibility to other diseases between the transgenic and conventional lines.”
Surprisingly, the application of glyphosate to the transgenic line actually increased yield compared to plants not treated with the herbicide. Yield measures included marketable ear number, marketable ear mass, and kernel mass.
“That was very interesting, because the study was maintained free of weeds,” says Williams. “Why would there be higher yields in glyphosate-treated plants when there were no weeds in the plots?”
The explanation may be hormesis, in which plant growth is stimulated as a result of a low, sub-lethal dose of a toxin. Although the mechanisms of hormesis have yet to be determined, similar effects have been observed in glyphosate-treated plants in other studies.
“One quart per acre of the glyphosate product would certainly be sub-lethal to a glyphosate resistant cultivar. Perhaps this dose stimulated crop growth.” Williams notes. “In any event, we know the transgene provided a similar level of resistance to glyphosate, as observed previously in other crops.”
In summary, the study showed neither an increased risk of Goss’s wilt nor a yield penalty with use of the GR+Bt transgene or glyphosate in sweet corn.

Fossil analysis pushes back human split from other primates by two million years
Posted by Acubiz | BlogTeam analysis of these 8-million-year-old Chororapithecus teeth fossils provided insights into the human-gorilla evolutionary split.
Credit: Gen Suwa
A paper in the latest issue of the journal Nature suggests a common ancestor of apes and humans,Chororapithecus abyssinicus, evolved in Africa, not Eurasia, two million years earlier than previously thought.
“Our new research supports early divergence: 10 million years ago for the human-gorilla split and 8 million years ago for our split from chimpanzees,” said Los Alamos National Laboratory geologist and senior team member Giday WoldeGabriel. “That’s at least 2 million years earlier than previous estimates, which were based on genetic science that lacked fossil evidence.”
“Our analysis of C. abyssinicus fossils reveals the ape to be only 8 million years old, younger than previously thought. This is the time period when human and African ape lines were thought to have split, but no fossils from this period had been found until now,” WoldeGabriel said.
Chimpanzees, gorillas, orangutans and humans compose the biological family Hominidae. Our knowledge of hominid evolution — that is, when and how humans evolved away from the great ape family tree — has significantly increased in recent years, aided by unearthed fossils from Ethiopia, including the C. abyssinicus, a species of great ape.
The international team that discovered the extinct gorilla-like species C. abyssinicus (reported in the journal Nature in 2007) reports new field observations and geological techniques that the authors say revise the age-constraint of the human split from their brethren.
The authors’ new paper, “New geological and palaeontological age constraint for the gorilla-human lineage split,” was published this week in Nature.WoldeGabriel coauthored the paper and his role was to characterize the volcanic ash and provide chemistry for local and regional correlation of the ashes sandwiching the fossils from Ethiopia’s Chorora area, a region where copious volcanic eruptions and earthquakes entombed fossils recently uplifted via ground motion and erosion.
Filling Gaps in the Fossil Record
Most of the senior members of the Chorora research team also belong to the Middle Awash project team that has recovered the fossil remains of at least eight hominid species, including some of the earliest hominids, spanning nearly 6 million years.
In the 1990s, before this team excavated the gorilla-like C. abyssinicus, they discovered the nearly intact skeleton of the 4.4-million-year-old speciesArdipithecus ramidus (nicknamed “Ardi”) and its relative, the million-year-older species Ardipithecus kadabba. These Ardipithecus fossils were the earliest ancestor of humans after they diverged from the main ape lineage of the primate family tree, neither ape-like nor chimp-like, yet not human either. Notably, both were bipedal — they walked upright.
While the team was still investigating Ardi and Kadabba, they published their results about C. abyssinicus. From the collection of nine fossilized teeth from multiple C. abyssinicus individuals, the team surmised that these teeth were gorilla-like, adapted for a fibrous diet. Based on their research from the Chorora, Kadabba and Ardi finds, the team says the common ancestor of chimps and humans lived earlier than had been evidenced by genetic and molecular studies, which placed the split about 5 million years ago.
According to the paper, C. abyssinicus revealed answers about gorilla lineage but also provided fossil evidence that our common ancestor migrated from Africa, not Eurasia, where fossils were more prolific prior to this discovery of multiple skeletons. While some skeptics say that more fossil evidence is needed before they accept this team’s conclusions, many agree that the discovery of a fossil ape from this time period is important since only one other had been found.
Extensive Analysis Provides New Evidence
WoldeGabriel and the research team used a variety of methods to determine the age of teeth they found at the Chorora Formation. They estimated the age of the volcanic rocks and sediments that encased the fossils with argon-dating and paleomagnetic methods. The team investigated patterns of magnetic reversals — another method to determine age based on knowledge about an era’s magnetic orientation — and calibrated the sediments containing the fossils using Geomagnetic Polarity Time Scale (GPTS).
Through fieldwork, volcanic ash chemistry and geochronology, WoldeGabriel helped nail down the age of the fossils to approximately 8 million years old. Based on this new fossil evidence and analysis, the team suggests that the human branch of the tree (shared with chimpanzees) split away from gorillas about 10 million years ago — at least 2 million years earlier than previously claimed.
Cytokine plays dual role in regulating inflammatory bowel disease, study finds
Posted by Acubiz | BlogSmall proteins that affect communication between cells play an important role in regulating inflammation that occurs during inflammatory bowel disease, according to researchers at Georgia State University, Emory University, the University of Michigan and Amgen, a biotechnology company.
The researchers compared immune cells in mice with and without intestinal inflammation and identified a new factor, a cytokine called IL-36, that is expressed in the inflamed intestine of mice. They explored the role of this cytokine to determine if it’s promoting disease, helping to protect against disease or simply associated with the disease. The findings are published inThe Journal of Immunology.
“What we found was quite striking,” said Tim Denning, lead author of the study and associate professor in the Institute for Biomedical Sciences at Georgia State. “If we block the effects of this cytokine, IL-36, in a mouse model of intestinal inflammation, the mice were better and had less disease early on, which suggested that this was a pro-inflammatory cytokine. However, when we assessed the ability of mice deficient in the receptor for IL-36 to heal, which is a vital part of resolving intestinal inflammation, they were completely unable to do so. The study highlights the important role of IL-36, not only in driving some of the inflammatory process, but also in helping to resolve the inflammation.”
Inflammatory bowel disease (IBD), chronic inflammation of all or part of the digestive tract, affects about 1.5 million Americans. The two main types, Crohn’s disease and ulcerative colitis, develop from uncontrolled inflammation in the intestine, which leads to severe diarrhea, pain, fatigue, weight loss and even death.
There are trillions of helpful bacteria inside human intestines, and the immune system is trained not to react aggressively. However, in people with IBD, the immune system doesn’t tolerate these bacteria and instead fights against them.
In this study, the researchers investigated the factors that may be regulating the immune system’s balance between tolerating these bacteria and reacting aggressively against them. They discovered the dual role of IL-36 in both promoting intestinal inflammation and resolving or healing that inflammation. The findings highlight the significance of understanding the timing and phase of disease, Denning said.
“Treatments that block certain factors, regardless of knowing the role it may be playing at a certain stage of disease, could lead to a poor outcome and may be the reason some clinical trials fail,” Denning said. “It is key to understand what phase of disease patients are in, what cytokines are expressed and the appropriate therapeutic targets during these distinct phases. Oftentimes, blocking a factor is not universally beneficial. Immune responses and inflammation, which are often viewed as deleterious, can be both good and bad depending on the context.”
For instance, blocking IL-36 may be beneficial in certain phases of the disease, but eliminating this factor could also make the body unable to recruit and activate cells necessary for healing and resolution of the disease.
Therefore, it’s important to understand the pathways IL-36 affects because this cytokine could be valuable in other stages of the disease, he said.
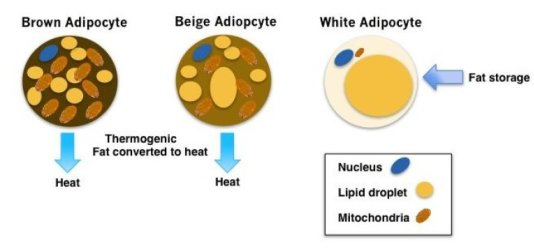
‘Beiging’ white fat cells to fight diabetes
Posted by Acubiz | BlogThe different types of fat cells are shown.
Credit: Cassie Tan , PhD, Perelman School of Medicine, University of Pennsylvania
Researchers are getting closer to learning how to turn white fat cells into brown fat cells, in a process called “beiging,” to bring down blood sugar levels and fight diabetes. The team, led by Joseph Baur, PhD, an assistant professor of Physiology in the Perelman School of Medicine at the University of Pennsylvania published their findings this month in the journal Diabetes.
“Beiging of white fat could be harnessed to fight diabetes by burning excess calories to cause a decrease in blood sugar,” Baur said. “Our work suggests that activation of the mTOR pathway plays a critical role in this process.” Induction of beige fat cells is considered a promising strategy to combat obesity because of this cell type’s ability to metabolize glucose and lipids, dissipating the resulting energy as heat.
Brown and white fat cells, or adipocytes, play different roles in the body. While white adipocytes store energy as large fat droplets, brown adipocytes contain smaller fat droplets and are specialized to burn fat to produce heat. To do this brown adipocytes are packed with the powerhouses called mitochondria that contain iron, which gives them their brown color. In fact, babies are born with brown fat along the upper back and shoulders to keep warm.
In adult humans, the recent discovery of brown fat “depots” is also associated with lower body weight. Brown-like fat cells, called beige adipocytes, also appear within white fat deposits in response to cold and other signals. The energy balance within the body is influenced by brown and beige adipocytes, which are stimulated into action by cold temperatures and other signals to burn fat and carbohydrates.
The primary tool used in these studies was rapamycin, a drug that inhibits the protein mTOR (mechanistic target of rapamycin), which can be found in two distinct protein complexes. It was first discovered as a byproduct of Streptomycin hygroscopicus, a bacterium found in a soil sample from Easter Island, an island also known as Rapa Nui, hence the name. Rapamycin is currently used as an immunosuppressant in organ transplant, but has recently attracted attention when it was discovered to extend lifespan in mice.
Interestingly, in 2012, Baur’s lab discovered that rapamycin also causes insulin resistance due to its ability to inhibit both arms of the mTOR signaling pathway controlled by the protein complexes mTORC1 and mTORC2. They showed in an animal model that these two arms could, in principle, be separated to dissect which pathway controls longevity versus endocrine effects.
In terms of physiology, mTOR signaling is involved in the control of blood sugar and cholesterol levels, and its inhibition increases the risk of diabetes. While previous studies suggested that mTORC1 inhibition would promote beiging of white fat cells, Baur’s present work supports the notion that mTORC1 activity is actually required for cold-induced beiging of white fat cells. If activating mTORC1 directly can bring about the same result, then this approach could potentially be applied to combat diabetes.
In the Diabetes study, the team shows that rapamycin blocks the ability of cold or drugs that activate a specific neurotransmitter pathway to induce the appearance of beige fat cells. Accordingly, rapamycin-treated mice are cold-intolerant and fail to maintain body temperature and weight when moved to a colder environment.
The findings demonstrate a positive role for mTORC1 in the recruitment of beige fat cells to white fat depots, which could explain some of the negative metabolic effects of mTOR inhibition.
“Our study highlights the complex interconnection between mTOR signaling and metabolism,” said first author Cassie Tran, PhD, a postdoctoral fellow in the Baur lab. “It will be critical in moving forward to determine the specific targets downstream of mTOR that are causing the negative metabolic effects in order to create better drugs and one day drugs that might also extend heathspan. The discovery of a critical signaling pathway for beige-fat formation also suggests the opportunity to target this pathway to therapeutically increase the number of heat-producing cells in obese or diabetic patients.”
Effectiveness of a herpesvirus CMV-based vaccine against Ebola
Posted by Acubiz | BlogThis study represents a crucial step in the translation of herpesvirus-based Ebola virus vaccines into humans and other great apes.
The current CMV vaccine was designed to make the Ebola virus GP at later times. (stock image)
Credit: © ChiccoDodiFC / Fotolia
As the latest in a series of studies, researchers at Plymouth University, National Institutes of Health and University of California, Riverside, have shown the ability of a vaccine vector based on a common herpesvirus called cytomegalovirus (CMV) expressing Ebola virus glycoprotein (GP), to provide protection against Ebola virus in the experimental rhesus macaque, non-human primate (NHP) model. Demonstration of protection in the NHP model is regarded as a critical step before translation of Ebola virus vaccines into humans and other great apes.
The study is published today, Monday 15th February, in the online journal from Nature publishing, Scientific Reports.
In addition to establishing the potential for CMV-based vaccines against Ebola virus, these results are exciting from the potential insight they give into the mechanism of protection. Herpesvirus-based vaccines can theoretically be made to produce their targeted protein (in this case, Ebola virus GP) at different times following vaccination. The current CMV vaccine was designed to make the Ebola virus GP at later times. This resulted in the surprising production of high levels of antibodies against Ebola virus with no detectable Ebola-specific T cells. This immunological shift towards antibodies has never been seen before for such primate herpesvirus-based vaccines, where responses are always associated with large T cell responses and poor to no antibodies.
“This finding was complete serendipity,” says Dr Michael Jarvis who is leading the project at Plymouth University. “Although we will definitely need to explore this finding further, it suggests that we may be able to bias immunity towards either antibodies or T cells based on the time of target antigen production. This is exciting not just for Ebola, but for vaccination against other infectious as well as non-infectious diseases.”
A largely untold story is the devastating effect Ebola virus is having on wild great ape populations in Africa. Although the present study administered the vaccine by direct inoculation, a CMV-based vaccine that can spread from animal to animal may be one approach to protect such inaccessible wild animal populations that are not amenable to vaccination by conventional approaches. The current study is a step forward, not only for conventional Ebola virus vaccines for use in humans, but also in the development of such ‘self-disseminating vaccines’ to target Ebola in great apes, and other emerging infectious diseases in their wild animal host before they fully establish themselves in humans.

Caught in the act: Astronomers find a rare supernova ‘impostor’ in a nearby galaxy
Posted by Acubiz | BlogThe galaxy NGC 300, home to the unusual system Binder and her colleagues studied. The spiral galaxy is over 6 million light years away.
Credit: NASA/JPL-Caltech/OCIW
Breanna Binder, a University of Washington postdoctoral researcher in the Department of Astronomy and lecturer in the School of STEM at UW Bothell, spends her days pondering X-rays.
As she and her colleagues report in a new paper published Feb. 12, 2016 in the Monthly Notices of the Royal Astronomical Society, they recently solved a mystery involving X-rays — a case of X-rays present when they shouldn’t have been. This mystery’s unusual main character — a star that is pretending to be a supernova — illustrates the importance of being in the right place at the right time.
Such was the case in May 2010 when an amateur South African astronomer pointed his telescope toward NGC300, a nearby galaxy. He discovered what appeared to be a supernova — a massive star ending its life in a blaze of glory.
“Most supernovae are visible for a short time and then — over a matter of weeks — fade from view,” said Binder.
After a star explodes as a supernova, it usually leaves behind either a black hole or what’s called a neutron star — the collapsed, high-density core of the former star. Neither should be visible to Earth after a few weeks. But this supernova — SN 2010da — still was.
“SN 2010da is what we call a ‘supernova impostor’ — something initially thought to be a supernova based on a bright emission of light, but later to be shown as a massive star that for some reason is showing this enormous flare of activity,” said Binder.
Many supernova impostors appear to be massive stars in a binary system — two stars in orbit of one another. Stellar astrophysicists think that the impostor’s occasional flare-ups might be due to perturbations from its neighbor.
For SN 2010da, the story appeared to be over until September 2010 — four months after it was confirmed as an impostor — when Binder pointed NASA’s Chandra X-ray Observatory toward NGC300 and found something unexpected.
“There was just this massive amount of X-rays coming from SN 2010da, which you should not see coming from a supernova impostor,” she said.
Binder considered a variety of explanations. For example, material from the star’s corona could be hitting a nearby dust cloud. But that would not produce the level of X-rays she had observed. Instead, the intensity of the X-rays coming from SN 2010da were consistent with a neutron star — the dense, collapsed core remnant of a supernovae.
“A neutron star at this location would be surprising,” said Binder, “since we already knew that this star was a supernova impostor — not an actual supernova.”
In 2014, Binder and her colleagues looked at this system again with Chandra and, for the first time, the Hubble Space Telescope. They found the impostor star and those puzzling X-ray emissions. Based on these new data, they concluded that, like many other supernovae impostors, SN 2010da likely has a companion. But, unlike any other supernovae impostor binary reported to date, SN 2010da is probably paired with a neutron star.
“If this star’s companion truly is a neutron star, that would mean that the neutron star was once a giant, massive star that underwent its own supernova explosion in the past,” said Binder. “The fact that this supernova event didn’t expel the other star, which is 20 to 25 times the mass of our sun, makes this an incredibly rare type of binary system.”
To understand how this unusual binary system could form, Binder and her colleagues considered the age of the stars in this region of space. Looking at stellar size and luminosity, they discovered that most nearby stars were created in two bursts — one 30 million years ago and the other less than 5 million years ago. But neither SN 2010da nor its presumed neutron star companion could’ve been created in the older burst of starbirth.
“Most stars that are as massive as these usually live 10 to 20 million years, not 30 million,” said Binder. “The most massive, hottest stars can form, grow, swell, explode and leave a neutron star emitting X-rays in about 5 million years.”
Surveys of the galaxy as recently as 2007 and 2008 detected no X-ray emissions from the location of SN 2010da. Instead, Binder believes that the X-rays they first found in 2010 represent the neutron star “turning on” for the first time after its formation. The X-rays are likely produced when material from the impostor star is transferred to the neutron star companion.
“That would mean that this is a really rare system at an early stage of formation,” said Binder, “and we could learn a lot about how massive stars form and die by continuing to study this unique pairing.”
One mystery solved, Binder would like to keep looking at SN 2010da, seeing what else she can learn about its formation and evolution. Its home galaxy, which has yielded unique pairings previously, is sure to keep her busy. She is also planning a follow-up study of other recent supernova impostors with the help of an undergraduate research assistant at UW Bothell.
Researchers create ‘mini-brains’ in lab to study neurological diseases
Posted by Acubiz | BlogUse of human-derived structures could allow for better research and reduce animal testing.
Researchers at the Johns Hopkins Bloomberg School of Public Health say they have developed tiny “mini-brains” made up of many of the neurons and cells of the human brain — and even some of its functionality — and which can be replicated on a large scale.
The researchers say that the creation of these “mini-brains,” which will be discussed at the American Association for the Advancement of Science conference in Washington, DC on Feb. 12 at a press briefing and in a session on Feb. 13, could dramatically change how new drugs are tested for effectiveness and safety, taking the place of the hundreds of thousands of animals used for neurological scientific research in the United States. Performing research using these three-dimensional “mini-brains” — balls of brain cells that grow and form brain-like structures on their own over the course of eight weeks — should be superior to studying mice and rats because they are derived from human cells instead of rodents, they say.
“Ninety-five percent of drugs that look promising when tested in animal models fail once they are tested in humans at great expense of time and money,” says study leader Thomas Hartung, MD, PhD, the Doerenkamp-Zbinden Professor and Chair for Evidence-based Toxicology at the Bloomberg School. “While rodent models have been useful, we are not 150-pound rats. And even though we are not balls of cells either, you can often get much better information from these balls of cells than from rodents.
“We believe that the future of brain research will include less reliance on animals, more reliance on human, cell-based models.”
Hartung and his colleagues created the brains using what are known as induced pluripotent stem cells (iPSCs). These are adult cells that have been genetically reprogrammed to an embryonic stem cell-like state and then are stimulated to grow into brain cells. Cells from the skin of several healthy adults were used to create the mini-brains, but Hartung says that cells from people with certain genetic traits or certain diseases can be used to create brains to study various types of pharmaceuticals. He says the brains can be used to study Alzheimer’s disease, Parkinson’s disease, multiple sclerosis and even autism. Projects to study viral infections, trauma and stroke have been started.
Hartung’s mini-brains are very small — at 350 micrometers in diameter, or about the size of the eye of a housefly, they are just visible to the human eye — and hundreds to thousands of exact copies can be produced in each batch. One hundred of them can grow easily in the same petri dish in the lab. After cultivating the mini-brains for about two months, the brains developed four types of neurons and two types of support cells: astrocytes and oligodendrocytes, the latter of which go on to create myelin, which insulates the neuron’s axons and allows them to communicate faster.
The researchers could watch the myelin developing and could see it begin to sheath the axons. The brains even showed spontaneous electrophysiological activity, which could be recorded with electrodes, similar to an electroencephalogram, also known as EEG. To test them, the researchers placed a mini-brain on an array of electrodes and listened to the spontaneous electrical communication of the neurons as test drugs were added.
“We don’t have the first brain model nor are we claiming to have the best one,” says Hartung, who also directs the School’s Center for Alternatives to Animal Testing.
“But this is the most standardized one. And when testing drugs, it is imperative that the cells being studied are as similar as possible to ensure the most comparable and accurate results.”
Hartung is applying for a patent for the mini-brains and is also developing a commercial entity called ORGANOME to produce them. He hopes production can begin in 2016. He says they are easily reproducible and hopes to see them used by scientists in as many labs as possible. “Only when we can have brain models like this in any lab at any time will we be able to replace animal testing on a large scale,” he says.
Association for the Advancement of Science conference in Washington, DC on Feb. 12 at a press briefing and in a session on Feb. 13, could dramatically change how new drugs are tested for effectiveness and safety, taking the place of the hundreds of thousands of animals used for neurological scientific research in the United States. Performing research using these three-dimensional “mini-brains” — balls of brain cells that grow and form brain-like structures on their own over the course of eight weeks — should be superior to studying mice and rats because they are derived from human cells instead of rodents, they say.
“Ninety-five percent of drugs that look promising when tested in animal models fail once they are tested in humans at great expense of time and money,” says study leader Thomas Hartung, MD, PhD, the Doerenkamp-Zbinden Professor and Chair for Evidence-based Toxicology at the Bloomberg School. “While rodent models have been useful, we are not 150-pound rats. And even though we are not balls of cells either, you can often get much better information from these balls of cells than from rodents.
“We believe that the future of brain research will include less reliance on animals, more reliance on human, cell-based models.”
Hartung and his colleagues created the brains using what are known as induced pluripotent stem cells (iPSCs). These are adult cells that have been genetically reprogrammed to an embryonic stem cell-like state and then are stimulated to grow into brain cells. Cells from the skin of several healthy adults were used to create the mini-brains, but Hartung says that cells from people with certain genetic traits or certain diseases can be used to create brains to study various types of pharmaceuticals. He says the brains can be used to study Alzheimer’s disease, Parkinson’s disease, multiple sclerosis and even autism. Projects to study viral infections, trauma and stroke have been started.
Hartung’s mini-brains are very small — at 350 micrometers in diameter, or about the size of the eye of a housefly, they are just visible to the human eye — and hundreds to thousands of exact copies can be produced in each batch. One hundred of them can grow easily in the same petri dish in the lab. After cultivating the mini-brains for about two months, the brains developed four types of neurons and two types of support cells: astrocytes and oligodendrocytes, the latter of which go on to create myelin, which insulates the neuron’s axons and allows them to communicate faster.
The researchers could watch the myelin developing and could see it begin to sheath the axons. The brains even showed spontaneous electrophysiological activity, which could be recorded with electrodes, similar to an electroencephalogram, also known as EEG. To test them, the researchers placed a mini-brain on an array of electrodes and listened to the spontaneous electrical communication of the neurons as test drugs were added.
“We don’t have the first brain model nor are we claiming to have the best one,” says Hartung, who also directs the School’s Center for Alternatives to Animal Testing.
“But this is the most standardized one. And when testing drugs, it is imperative that the cells being studied are as similar as possible to ensure the most comparable and accurate results.”
Hartung is applying for a patent for the mini-brains and is also developing a commercial entity called ORGANOME to produce them. He hopes production can begin in 2016. He says they are easily reproducible and hopes to see them used by scientists in as many labs as possible. “Only when we can have brain models like this in any lab at any time will we be able to replace animal testing on a large scale,” he says.